Abstract
Background: Cannabinoid compounds have been approved as prescription drugs for treating various human ailments. However, the production using both microbial and plant-based sources is time-consuming and expensive because their yield is extremely low. Tetraketide synthase (TKS), the key enzyme in the biosynthesis of cannabinoid compounds, produces only 4% of the intermediate compound olivetolic acid. However, it may be possible to rearrange the carbon metabolic flux of TKS using genetic methods to increase the overall yields of cannabinoid compounds. In this context, protein engineering is an economically beneficial and viable solution to improve the catalytic activity of TKS. However, the ability to produce enzyme variants significantly exceeds the capacity to screen and identify high producers, creating a bottleneck in the enzyme engineering process.
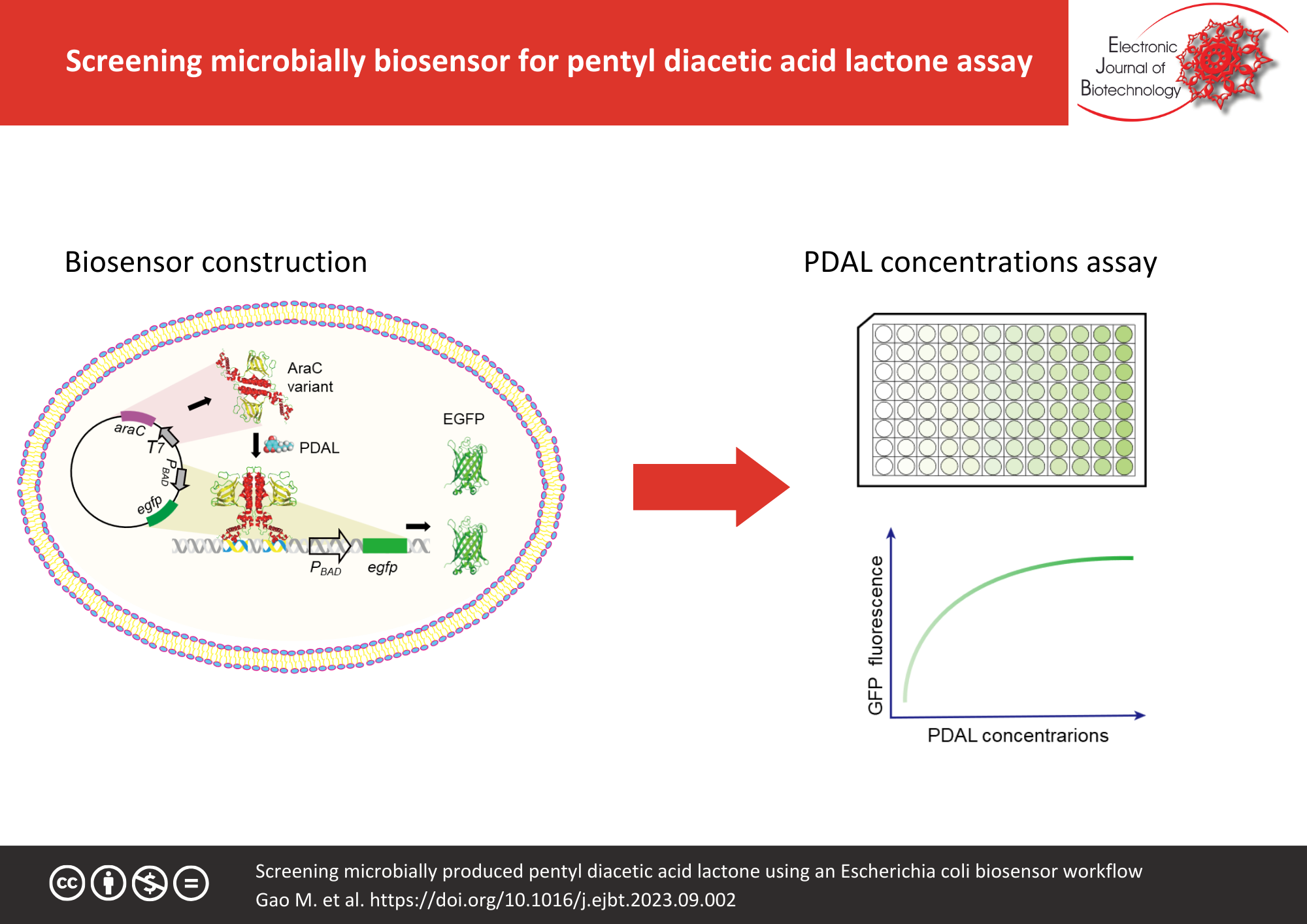
Results: This study constructed an Escherichia coli-based biosensor workflow for detecting the byproduct pentyl diacetic acid lactone (PDAL). Rational design was used to generate E. coli strains with mutant regulatory protein AraC and an altered effector PDAL to control the transcription of gfp and kanamycin. The developed biosensor could detect PDAL at the concentrations of the operational range from microbial cell culture and cell-free catalytic system.
Conclusions: The E. coli-based biosensor developed in this study efficiently detected PDAL with high throughput and low cost.
References
Tang XY, K Eitel, L Kaysser, et al. A two-step sulfation in antibiotic biosynthesis requires a type III polyketide synthase. Nat Chem Biol, 2013;9(10):610-615. https://doi.org/10.1038/nchembio.1310 PMid: 23912167
Bisht R, A Bhattacharyya, A Shrivastava, et al. An overview of the medicinally important plant type III PKS derived polyketides. Front Plant Sci, 2021;12:746908. https://doi.org/10.3389/fpls.2021.746908 PMid: 34721474
Nivina A, SH Paredes, HB Fraser, et al. GRINS: Genetic elements that recode assembly-line polyketide synthases and accelerate their diversification. Proc Natl Acad Sci USA, 2021;118(26):e2100751118. https://doi.org/10.1073/pnas.2100751118 PMid: 34162709
Zhou YC, WT Tao, Z Qi, et al. Structural and mechanistic insights into chain release of the polyene PKS thioesterase domain. Acs Catal, 2022;12(1):762-776. https://doi.org/10.1021/acscatal.1c04991
Brauer A, QQ Zhou, GLC Grammbitter, et al. Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis. Nat Chem, 2020;12(8):755-763. https://doi.org/10.1038/s41557-020-0491-7 PMid: 32632186
Sulpizio A, CEW Crawford, RS Koweek, et al. Probing the structure and function of acyl carrier proteins to unlock the strategic redesign of type II polyketide biosynthetic pathways. J Biol Chem, 2021;296:100328. https://doi.org/10.1016/j.jbc.2021.100328 PMid: 33493513
Walker PD, ANM Weir, CL Willis, et al. Polyketide ?-branching: Diversity, mechanism and selectivity. Nat Prod Rep, 2021;38(4):723-756. https://doi.org/10.1039/D0NP00045K PMid: 33057534
Guo DM, HY Wang, SM Zhang, et al. The type III polyketide synthase supergene family in plants: Complex evolutionary history and functional divergence. Plant J, 2022;112(2):414-428. https://doi.org/10.1111/tpj.15953 PMid: 36004534
Wang XH, BW Gao, Y Nakashima, et al. Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood. Nat Commun, 2022;13(1):348. https://doi.org/10.1038/s41467-022-27971-z PMid: 35039506
Zargar A, R Lal, L Valencia, et al. Chemoinformatic-guided engineering of polyketide synthases. J Am Chem Soc, 2020;142(22):9896-9901. https://doi.org/10.1021/jacs.0c02549 PMid: 32412752
Helfrich EJN, R Ueoka, MG Chevrette, et al. Evolution of combinatorial diversity in trans-acyltransferase polyketide synthase assembly lines across bacteria. Nat Commun, 2021;12(1):1422. https://doi.org/10.1038/s41467-021-21163-x PMid: 33658492
Kalkreuter E, KS Bingham, AM Keeler, et al. Computationally-guided exchange of substrate selectivity motifs in a modular polyketide synthase acyltransferase. Nat Commun, 2021;12(1):2193. https://doi.org/10.1038/s41467-021-22497-2 PMid: 33850151
He LM, R Zhang, JD Shen, et al. Improving the low-temperature properties of an exo-inulinase via the deletion of a loop fragment located in its catalytic pocket. Electron J Biotechnol, 2022;55:1-8. https://doi.org/10.1016/j.ejbt.2021.09.004
Radwan MM, S Chandra, S Gul, et al. Cannabinoids, phenolics, terpenes and alkaloids of Cannabis. Molecules, 2021;26(9):2774. https://doi.org/10.3390/molecules26092774 PMid: 34066753
Hryhorowicz S, M Kaczmarek-Rys, A Zielinska, et al. Endocannabinoid system as a promising therapeutic target in inflammatory bowel disease - a systematic review. Front Immunol, 2021;12:790803. https://doi.org/10.3389/fimmu.2021.790803 PMid: 35003109
Sledzinski P, J Zeyland, R Slomski, et al. The current state and future perspectives of cannabinoids in cancer biology. Cancer Med, 2018;7(3):765-775. https://doi.org/10.1002/cam4.1312 PMid: 29473338
Blasco-Benito S, E Moreno, M Seijo-Vila, et al. Therapeutic targeting of HER2-CB2R heteromers in HER2-positive breast cancer. Proc Natl Acad Sci USA, 2019;116(9):3863-3872. https://doi.org/10.1073/pnas.1815034116 PMid: 30733293
Andradas C, A Truong, J Byrne, et al. The role of cannabinoids as anticancer agents in pediatric oncology. Cancers, 2021;13(1):157. https://doi.org/10.3390/cancers13010157 PMid: 33466435
Marz?da P, M Drozd, P Wróblewska-?uczka, et al. Cannabinoids and their derivatives in struggle against melanoma. Pharmacol Rep, 2021;73(6):1485-1496. https://doi.org/10.1007/s43440-021-00308-1 PMid: 34264513
Villalobos P, MI Chavez, Y Olguin, et al. The application of polymerized lipid vesicles as colorimetric biosensors for real-time detection of pathogens in drinking water. Electron J Biotechnol, 2012;15(1). https://doi.org/10.2225/vol15-issue1-fulltext-5
Villalonga A, A Sanchez, B Mayol, et al. Electrochemical biosensors for food bioprocess monitoring. Curr Opin Food Sci, 2022;43:18-26. https://doi.org/10.1016/j.cofs.2021.09.006
Farina D, M Zinellu, M Fanari, et al. Development of a biosensor telemetry system for monitoring fermentation in craft breweries. Food Chem, 2017;218:479-486. https://doi.org/10.1016/j.foodchem.2016.09.092 PMid: 27719939
Ejeian F, P Etedali, H-A Mansouri-Tehrani, et al. Biosensors for wastewater monitoring: A review. Biosens Bioelectron, 2018;118:66-79. https://doi.org/10.1016/j.bios.2018.07.019 PMid: 30056302
Gavrilas S, CS Ursachi, S Perta-Crisan, et al. Recent trends in biosensors for environmental quality monitoring. Sensors, 2022;22(4):1513. https://doi.org/10.3390/s22041513 PMid: 35214408
Eyvazi S, B Baradaran, A Mokhtarzadeh, et al. Recent advances on development of portable biosensors for monitoring of biological contaminants in foods. Trends Food Sci Tech, 2021;114:712-721. https://doi.org/10.1016/j.tifs.2021.06.024
Abid SA, AA Muneer, IMS Al-Kadmy, et al. Biosensors as a future diagnostic approach for COVID-19. Life Sci, 2021;273:119117. https://doi.org/10.1016/j.lfs.2021.119117 PMid: 33508293
Tang S-Y, PC Cirino. Design and application of a mevalonate-responsive regulatory protein. Angew Chem Int Edit, 2011;50(5):1084-1086. https://doi.org/10.1002/anie.201006083 PMid: 21268200
Tang SY, S Qian, O Akinterinwa, et al. Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter. J Am Chem Soc, 2013;135(27):10099-10103. https://doi.org/10.1021/ja402654z PMid: 23786422
Tang S-Y, H Fazelinia, PC Cirino. AraC regulatory protein mutants with altered effector specificity. J Am Chem Soc, 2008;130(15):5267-5271. https://doi.org/10.1021/ja7109053 PMid: 18355019
Durrant JD, CAF de Oliveira, JA McCammon. POVME: An algorithm for measuring binding-pocket volumes. J Mol Graph Model, 2011;29(5):773-776. https://doi.org/10.1016/j.jmgm.2010.10.007 PMid: 21147010
Fricke PM, T Link, J Gatgens, et al. A tunablel-arabinose-inducible expression plasmid for the acetic acid bacterium Gluconobacter oxydans. Appl Microbiol Biotechnol, 2020;104(21):9267-9282. https://doi.org/10.1007/s00253-020-10905-4 PMid: 32974745
Duarte JM, I Barbier, Y Schaerli. Bacterial microcolonies in gel beads for high-throughput screening of libraries in synthetic biology. Acs Synth Biol, 2017;6(11):1988-1995. https://doi.org/10.1021/acssynbio.7b00111 PMid: 28803463
Gagne SJ, JM Stout, E Liu, et al. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc Natl Acad Sci USA, 2012;109(31):12811-12816. https://doi.org/10.1073/pnas.1200330109 PMid: 22802619
Chen W, S Zhang, P Jiang, et al. Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis. Metab Eng, 2015;30:149-155. https://doi.org/10.1016/j.ymben.2015.05.004 PMid: 26051748
Castillo-Hair SM, M Fujita, OA Igoshin, et al. An engineered B. subtilis inducible promoter system with over 10 000-fold dynamic range. Acs Synth Biol, 2019;8(7):1673-1678. https://doi.org/10.1021/acssynbio.8b00469 PMid: 31181163

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright (c) 2023 Electronic Journal of Biotechnology